

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
3D GREIT Reconstruction with a non-extruded volumeForward model with 3 rings of 12 electrodes
ellipsoid.center = [0;0;3];
ellipsoid.axis_a = [2;0;0];
ellipsoid.axis_b = [0;1;0];
ellipsoid.axis_c = [0;0;3.5];
ortho_brick.opposite_corner_a = [2; 1; 0];
ortho_brick.opposite_corner_b = [-2;-1; 3];
bg.intersection.ellipsoid = ellipsoid;
bg.intersection.ortho_brick = ortho_brick;
% shape
mdl = ng_mk_geometric_models(bg);
% add electrodes
fmdl = place_elec_on_surf(mdl,[12,0, 1.25, 1.75, 2.25],[0.1, 0, 0.02]);
% re-order electrodes and add stimulation
v = reshape(1:36,[],3)';
fmdl.electrode = fmdl.electrode(v(:));
fmdl.stimulation = mk_stim_patterns(36,1,[0 9],[0,5],{},1);
show_fem(fmdl);
print_convert GREIT3D_arbitrary01.jpg
Simulate dataimg = mk_image(fmdl, 1); vh = fwd_solve(img); esp = elem_select(fmdl, '(x-0.5).^2+(y-0.3).^2+(z-1.5).^2<0.2^2'); esn = elem_select(fmdl, '(x+0.5).^2+(y+0.3).^2+(z-2).^2<0.2^2'); img.elem_data = 1 + 0.01*esp - 0.01*esn; vi = fwd_solve(img); clf show_fem(img) print_convert GREIT3D_arbitrary_02a.jpg clf show_3d_slices(img,[1.5, 2],[-.5 .5],0) % cuts through the targets print_convert GREIT3D_arbitrary_02b.jpg clf img.fwd_model.mdl_slice_mapper.npx = 64; img.fwd_model.mdl_slice_mapper.npy = 32; show_slices(img,5) print_convert GREIT3D_arbitrary_02c.png 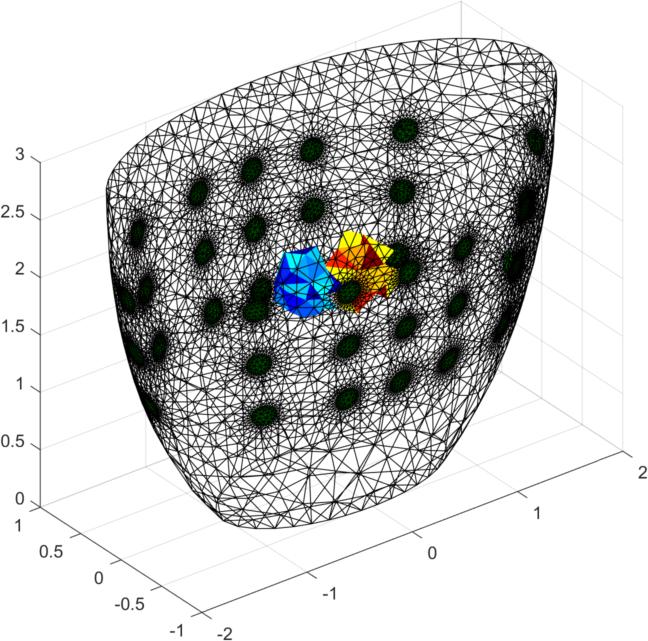
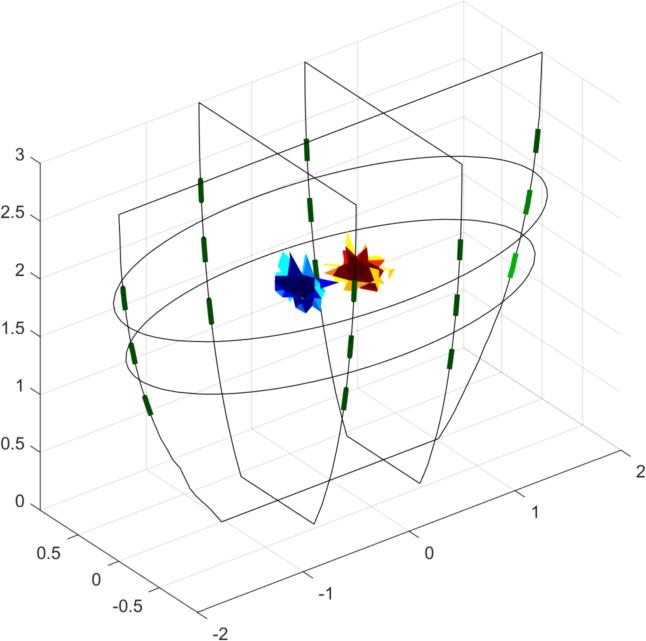
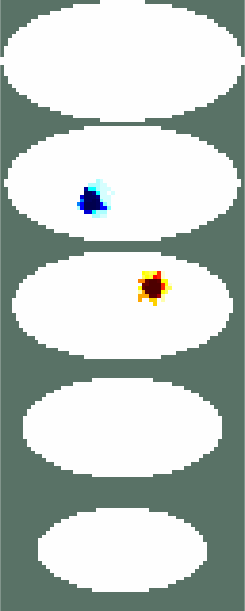
Figure: Three different views of the simulation model with targets Create reconstruction modelCreate a 3D voxel model to reconstruct on. Note that we commit the inverse crime by using the same forward model for simulation and reconstruction.% Setup a 3D rec_model vopt.cube_voxels = true; %vopt.imgsz = [64, 64, 64]; % looks nice, but takes very long vopt.imgsz = [48, 48, 48]; % A bit faster vopt.downsample = [2, 1]; % reduce density of targets vopt.save_memory = 1; [imdl, distr] = GREIT3D_distribution(fmdl,vopt); % limit training points to where the electrodes are distr(:,(distr(3,:)>2.5)) = []; distr(:,distr(3,:)<1) = []; gopt.distr = distr; gopt.noise_figure = 1.0; % train GREIT imdl3 = mk_GREIT_model(imdl, 0.2, [], gopt); Reconstruct and show images% reconstruct rimg = inv_solve(imdl3, vh, vi); clf show_fem(rimg) print_convert GREIT3D_arbitrary_04a.jpg clf show_3d_slices(rimg,[1.5, 2],[-.5 .5],0) % cuts through the targets print_convert GREIT3D_arbitrary_04b.jpg clf show_slices(rimg,5) print_convert GREIT3D_arbitrary_04c.png 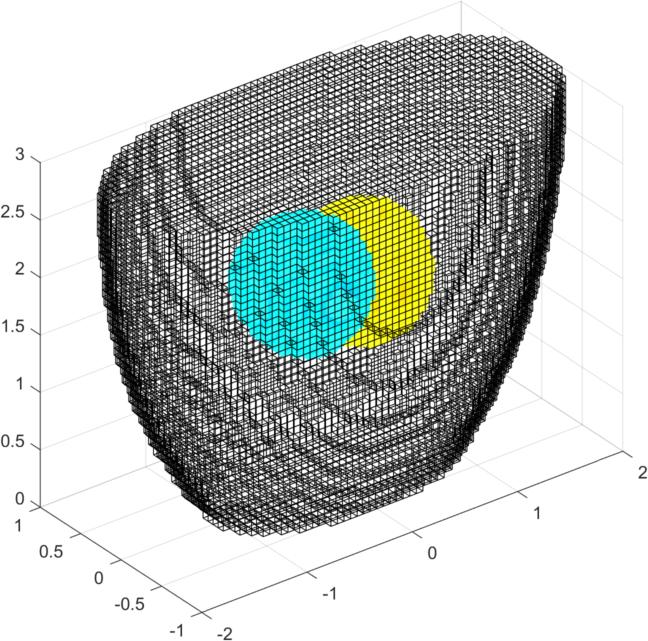
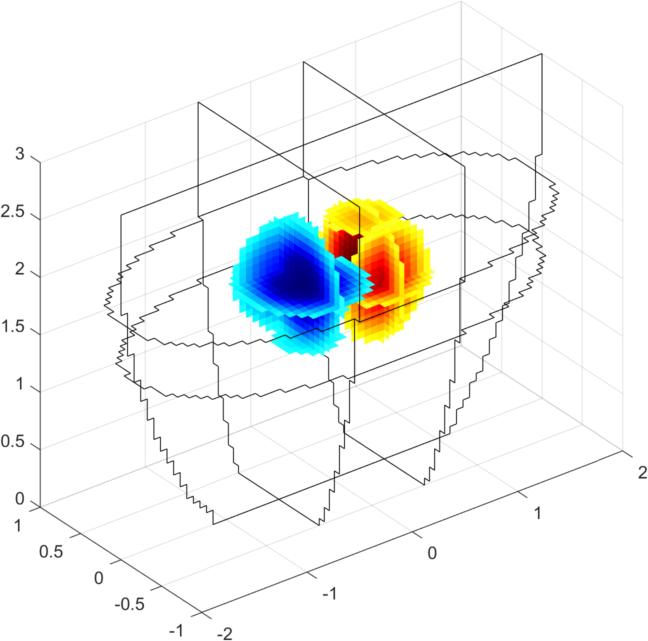
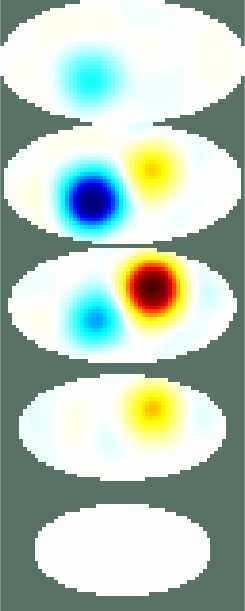
Figure: Three different views of the reconstruction image. Last Modified: $Date: 2024-04-20 15:44:10 -0400 (Sat, 20 Apr 2024) $ by $Author: aadler $ |