

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Algorithm PerformanceEIDORS provides an interface to test algorithm performance against the parameters defined for the GREIT algorithm: Amplitude (AR), Position Error (PE), Resolution (RES), Shape Deformation (SD), and Ringing (RNG)
% Test algorithm performance $Id: algorithm_performance01.m 2240 2010-07-04 14:41:32Z aadler $
% Reconstruct GREIT Images
imdl_gr = mk_common_gridmdl('GREITc1');
% Reconstruct backprojection Images
imdl_bp = mk_common_gridmdl('backproj');
% Reconstruct GN Images
imdl_gn = select_imdl( mk_common_model('d2c2', 16), {'Basic GN dif','Choose NF=0.5'});
test_performance( { imdl_gr, imdl_bp, imdl_gn } );
print_convert 'algorithm_performance01a.png' '-density 100'
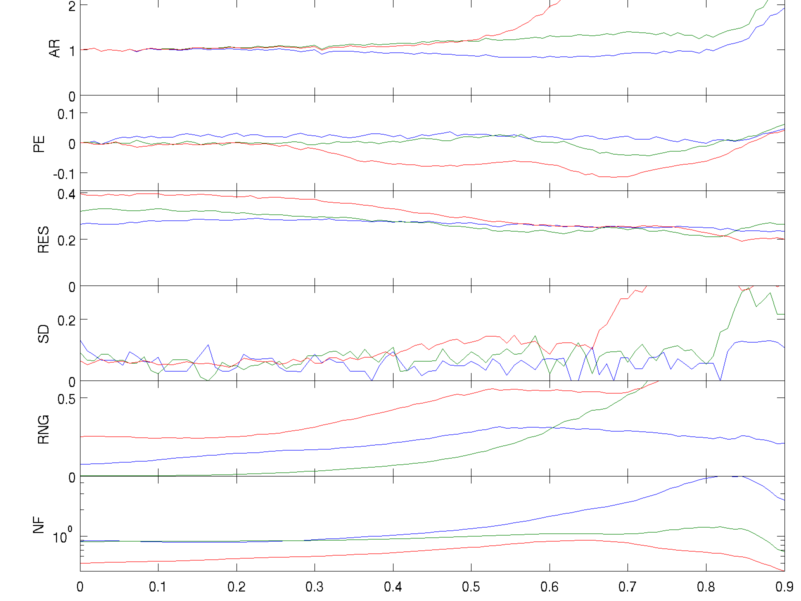
Figure: Performance of algorithms: blue: GREIT (v1) green: Sheffield backprojection red: One step Gauss Newton GREIT Test Parameters for different algorithmsThe GREIT figure of merit parameters to evaluate the performace of other algorithms.Simulate 3D object
% create 3D forward model
fmdl = ng_mk_cyl_models([2,1,0.08],[8,0.8,1.2],[0.05]);
fmdl.stimulation = mk_stim_patterns(16,1,[0,1],[0,1],{},1);
imgs= mk_image( fmdl, 1);
show_fem(imgs);
print_convert test_params01a.png '-density 50'

Figure: Simulation mesh Simulate target positionsr = 0.05; % target radius Npos = 20; % number of positions Xpos = linspace(0,0.9,Npos); % positions to simulated along x-axis Ypos = zeros(1,Npos); Zpos = ones(1,Npos); %% for off-plane, adjust the level (*1.5) xyzr = [Xpos; Ypos; Zpos; r*ones(1,Npos)]; [vh,vi] = simulate_movement(imgs, xyzr); Gauss-Newton Reconstruction Matrix
% 3D inverse model
imdl= mk_common_model('n3r2',[16,2]);
mdl= ng_mk_cyl_models([2,1,0.1],[8,0.8,1.2],[0.05]);
mdl.stimulation = mk_stim_patterns(16,1,[0,1],[0,1],{},1);
imdl.fwd_model = mdl;
img= mk_image( mdl, 1); J = calc_jacobian(img);
%% inverse solution (faster solution)
hp = 0.015;
RtR = prior_noser( imdl ); P= inv(RtR);
Rn = speye( size(J,1) );
imdl.solve = @solve_use_matrix;
imdl.solve_use_matrix.RM = P*J'/(J*P*J' + hp^2*Rn);
imgr= inv_solve(imdl,vh,vi);
Calculate and show GREIT parameters
%% calculate the GREIT parameters
levels =[inf,inf,Zpos(1)];
show_slices(imgr, levels);
imgr.calc_colours.npoints = 128;
imgr.calc_slices.levels=levels;
params = eval_GREIT_fig_merit(imgr, xyzr);
p_names = {'AR','PE','RES','SD','RNG'};
for i=1:5; subplot(5,1,i);
plot(params(i,:)); ylabel(p_names{i});
end
print_convert test_params04a.png '-density 100'

Figure: GREIT figure of merit parameters for GN algorithm |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $