

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Compare selection of hyperparameters for Total VariationTotal Variation typically uses a GN step to initialize the first iteration. We thus have a hyperparameter for the GN step (α1), and another for the GN interations (α2).Simluation objectSimulate shape with edges
% TV: Create object $Id: TV_hyperparams01.m 3365 2012-07-02 08:05:40Z aadler $
imb= mk_common_model('c2c2',16);
img= mk_image(imb.fwd_model, 1);
vh= fwd_solve( img );
img.elem_data([25,37,49:50,65:66,81:83,101:103,121:124])=2;
img.elem_data([95,98:100,79,80,76,63,64,60,48,45,36,33,22])=2;
vi= fwd_solve( img );
subplot(221)
show_fem(img);
axis equal; axis off;
print_convert TV_hyperparams01a.png '-density 100';

Figure: Shape that is to be reconstructed (generated on 576 element mesh) Create TV reconstruction model
% TV: Reconstruction model $Id: TV_hyperparams02.m 3428 2012-07-02 20:56:41Z bgrychtol $
maxit=20; % max number of iterations
imdl=mk_common_model('b2c0',16);
invtv= eidors_obj('inv_model', 'EIT inverse');
invtv.reconst_type= 'difference';
invtv.jacobian_bkgnd.value= 1;
invtv.fwd_model= imdl.fwd_model;
invtv.solve= @inv_solve_TV_pdipm;
invtv.R_prior= @prior_TV;
invtv.parameters.term_tolerance= 1e-6;
invtv.parameters.keep_iterations= 1;
invtv.parameters.max_iterations= maxit;
subplot(221)
show_fem(invtv.fwd_model);
axis equal; axis off;
print_convert TV_hyperparams02a.png '-density 100';
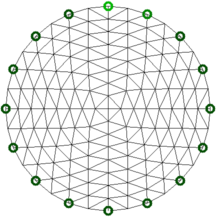
Figure: Reconstruction model (256 elements) Reconstruction with no noise
% Iterate over params $Id: TV_hyperparams03.m 3428 2012-07-02 20:56:41Z bgrychtol $
name_base = 'tv_hp_00_img-a1=%3.1f-a2=%3.1f.jpg';
alpha1list= [1.5:0.5:4.5];
alpha2list= [3.5:1.0:9.5];
for alpha2= alpha2list
for alpha1= alpha1list
invtv.hyperparameter.value = 10^-alpha2;
invtv.inv_solve_TV_pdipm.alpha1= 10^-alpha1;
imgs= inv_solve(invtv,vh,vi);
imgs.elem_data= imgs.elem_data(:,[1,2,5,maxit]);
imgs.calc_colours.window_range=.2;
imgs.calc_colours.clim=1.2;
out_img= show_slices(imgs);
imwrite(out_img, colormap, sprintf(name_base,alpha1,alpha2) );
end
end
To display these results, create an html table with matlab
% Generate HTML frame to view $Id: TV_hyperparams04.m 2639 2011-07-12 07:39:06Z bgrychtol $
fid= fopen('TV-params-NSR=0.html','w');
a=sprintf('%calpha;',38); % alpha
m=sprintf('%cminus;',38); % alpha
s=sprintf('%c',60); % less than
e=sprintf('%c',62); % greater than
tr= [s,'TR',e]; etr= [s,'/TR',e];
th= [s,'TH',e]; eth= [s,'/TH',e];
td= [s,'TD',e]; etd= [s,'/TD',e];
sub=[s,'SUB',e];esub=[s,'/SUB',e];
sup=[s,'SUP',e];esup=[s,'/SUP',e];
fprintf(fid,[s,'TABLE',e,tr,th]);
for alpha1= alpha1list
fprintf(fid,[th,a,sub,'1',esub,'=10',sup,m,'%3.1f',esup],alpha1);
end
fprintf(fid,'\n');
for alpha2= alpha2list
fprintf(fid,['\n',tr,'\n',th,a,sub,'2',esub,'=10', ...
sup,m,'%3.1f',esup,'\n'],alpha2);
for alpha1= alpha1list
fprintf(fid,[td,s,'img src="%s"',e,'\n'], sprintf(name_base,alpha1,alpha2) );
end
end
fprintf(fid,['\n',s,'/TABLE',e,'\n']);
fclose(fid);
Reconstruction Results (No Noise) (GN param = α1, TV param= α2)
Reconstruction with 20db SNR noiseAdd Noise % Add noise $Id: TV_hyperparams05.m 1535 2008-07-26 15:36:27Z aadler $ sig= sqrt(norm(vi.meas - vh.meas)); m= size(vi.meas,1); vi.meas = vi.meas + .01*sig*randn(m,1); Calculations
% Iterate over params $Id: TV_hyperparams06.m 3428 2012-07-02 20:56:41Z bgrychtol $
name_base= 'tv_hp_20_img-a1=%3.1f-a2=%3.1f.jpg';
alpha1list= [1.5:0.5:4.5];
alpha2list= [3.5:1.0:9.5];
for alpha2= alpha2list
for alpha1= alpha1list
invtv.hyperparameter.value = 10^-alpha2;
invtv.inv_solve_TV_pdipm.alpha1= 10^-alpha1;
imgs= inv_solve(invtv,vh,vi);
imgs.elem_data= imgs.elem_data(:,[1,2,5,maxit]);
imgs.calc_colours.window_range=.2;
imgs.calc_colours.clim=1.2;
out_img= show_slices(imgs);
imwrite(out_img, colormap, sprintf(name_base,alpha1,alpha2) );
end
end
To display these results, create an html table with matlab
% Generate HTML frame to view $Id: TV_hyperparams07.m 2733 2011-07-13 21:04:58Z anon-eidors $
fid= fopen('TV-params-NSR=0.01.html','w');
a=sprintf('%calpha;',38); % alpha
m=sprintf('%cminus;',38); % alpha
s=sprintf('%c',60); % less than
e=sprintf('%c',62); % greater than
tr= [s,'TR',e]; etr= [s,'/TR',e];
th= [s,'TH',e]; eth= [s,'/TH',e];
td= [s,'TD',e]; etd= [s,'/TD',e];
sub=[s,'SUB',e];esub=[s,'/SUB',e];
sup=[s,'SUP',e];esup=[s,'/SUP',e];
fprintf(fid,[s,'TABLE',e,tr,th]);
for alpha1= alpha1list
fprintf(fid,[th,a,sub,'1',esub,'=10',sup,m,'%3.1f',esup],alpha1);
end
fprintf(fid,'\n');
for alpha2= alpha2list
fprintf(fid,['\n',tr,'\n',th,a,sub,'2',esub,'=10', ...
sup,m,'%3.1f',esup,'\n'],alpha2);
for alpha1= alpha1list
fprintf(fid,[td,s,'img src="%s"',e,'\n'], sprintf(name_base,alpha1,alpha2) );
end
end
fprintf(fid,['\n',s,'/TABLE',e,'\n']);
fclose(fid);
Reconstruction Results (20dB SNR) (GN param = α1, TV param= α2)
|
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $