

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Simulate a moving 2D target on a fixed meshAnother way to simulate a moving target is to remesh around each new target. Doing this with distmesh involves directly accessing the mesh creation functions.Step 1: Create fine mesh (300 nodes)
% create model $Id: simulate_move2_01.m 6447 2022-12-02 12:16:51Z aadler $
% Model parameters
n_elec= 16;
n_nodes= 300;
% Create electrodes
refine_level=4; %electrode refinement level
elec_width= .1;
z_contact = 0.01;
% elect positions
th=linspace(0,2*pi,n_elec(1)+1)';th(end)=[];
elec_posn= [sin(th),cos(th)];
[elec_nodes, refine_nodes] = dm_mk_elec_nodes( elec_posn, ...
elec_width, refine_level);
% Define circular medium
fd=@(p) sum(p.^2,2)-1;
bbox = [-1,-1;1,1];
smdl= dm_mk_fwd_model( fd, [], n_nodes, bbox, ...
elec_nodes, refine_nodes, z_contact);
Step 2: Simulate movementTo simulate a target, we insert fixed note positions surrounding the target. Then using interp_mesh.m, we find the elements that are in the target position.
% create model $Id: simulate_move2_02.m 2781 2011-07-14 21:06:54Z bgrychtol $
% Create an object within the model
trg_ctr= [.7,.1];
trg_rad= 0.1;
th=linspace(0,2*pi,20)';th(end)=[];
trg_refine_nodes= [refine_nodes; ...
trg_rad*sin(th)+trg_ctr(1), ...
trg_rad*cos(th)+trg_ctr(2) ];
tmdl= dm_mk_fwd_model( fd, [], n_nodes, bbox, elec_nodes, ...
trg_refine_nodes, z_contact);
% find elements in size target
mdl_pts = interp_mesh( tmdl );
ctr_pts = mdl_pts - ones(size(mdl_pts,1),1)*trg_ctr;
in_trg =(sum(ctr_pts.^2,2) < trg_rad^2);
timg= eidors_obj('image','','fwd_model',tmdl,...
'elem_data',1 + in_trg*.5);
% Show output - full size
subplot(121); show_fem( smdl ); axis equal; axis([-1.1 1.1 -1.1 1.1]);
subplot(122); show_fem( timg ); axis equal; axis([-1.1 1.1 -1.1 1.1]);
print_convert simulate_move2_02a.png '-density 125'
% Show output - full size
subplot(121); show_fem( smdl ); axis equal; axis([.5,1.05,-.1,.3]);
subplot(122); show_fem( timg ); axis equal; axis([.5,1.05,-.1,.3]);
print_convert simulate_move2_02b.png '-density 125'

Figure: Left Simulation mesh homogeneous mesh Right Simulation mesh with target at (0.1,0.7) 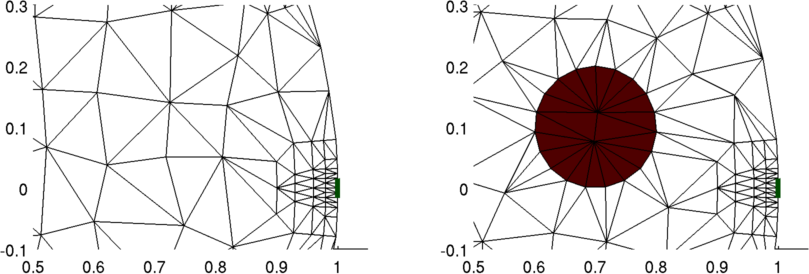
Figure: Magnification of areas of interest in the previous figure. Left Simulation mesh homogeneous mesh Right Simulation mesh with target at (0.1,0.7) Step 3: Simulate Homogeneous measurements
% simulate homogeneous $Id: simulate_move2_03.m 3273 2012-06-30 18:00:35Z aadler $
% stimulation pattern: adjacent
stim_pat= mk_stim_patterns(n_elec,1,'{ad}','{ad}',{},1);
smdl.stimulation= stim_pat;
himg= mk_image(smdl, 1);
vh= fwd_solve(himg);
Step 4: Simulate moving target and simulate measurements
% moving target $Id: simulate_move2_04.m 3273 2012-06-30 18:00:35Z aadler $
% Create a moving object within the model
trg_rad= 0.1;
radius= 0.75;
n_sims= 20;
contrast= 0.1;
clear vi;
for i= 1:n_sims;
thc= 2*pi*(i-1)/n_sims;
trg_ctr= radius*[cos(thc),sin(thc)];
trg_refine_nodes= [refine_nodes; ...
trg_rad*sin(th)+trg_ctr(1), ...
trg_rad*cos(th)+trg_ctr(2) ];
tmdl= dm_mk_fwd_model( fd, [], n_nodes, bbox, elec_nodes, ...
trg_refine_nodes, z_contact);
tmdl.stimulation = stim_pat;
% find elements in size target
mdl_pts = interp_mesh( tmdl );
ctr_pts = mdl_pts - ones(size(mdl_pts,1),1)*trg_ctr;
in_trg =(sum(ctr_pts.^2,2) < trg_rad^2);
% Create target image object
timg= mk_image(tmdl,1 + in_trg*contrast);
clf; show_fem(timg); axis equal
print_convert(sprintf('simulate_move2_04a%02d.png',i),'-density 50');
vi(i)= fwd_solve(timg);
end
In order to animate these meshes, we call the
imagemagick
convert function to create the animations
% Create animated graphics $Id: simulate_move2_05.m 6448 2022-12-02 12:19:15Z aadler $
% Trim images
system('find -name "simulate_move2_04a*.png" -exec convert -trim "{}" PNG8:"{}" ";"')
% Convert to animated Gif
system('convert -delay 50 simulate_move2_04a*.png -loop 0 simulate_move2_05a.gif')
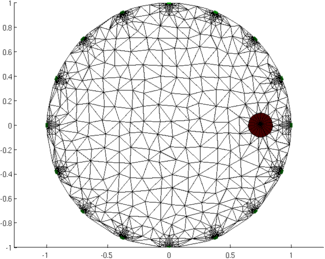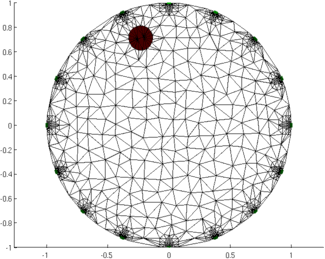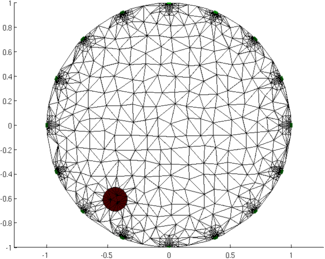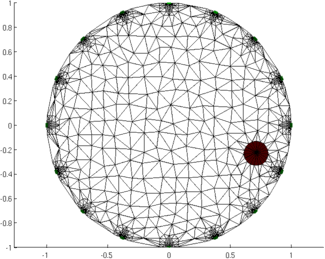
Figure: Animation of the simulation mesh surrounding a moving target Step 5: Reconstruct images
% Reconstruct images $Id: simulate_move2_06.m 2781 2011-07-14 21:06:54Z bgrychtol $
imdl= mk_common_model('c2c2',16);
img= inv_solve(imdl, vh, vi);
subplot(121); show_fem(img); axis image
subplot(122); show_slices(img)
print_convert simulate_move2_06a.png '-density 125'
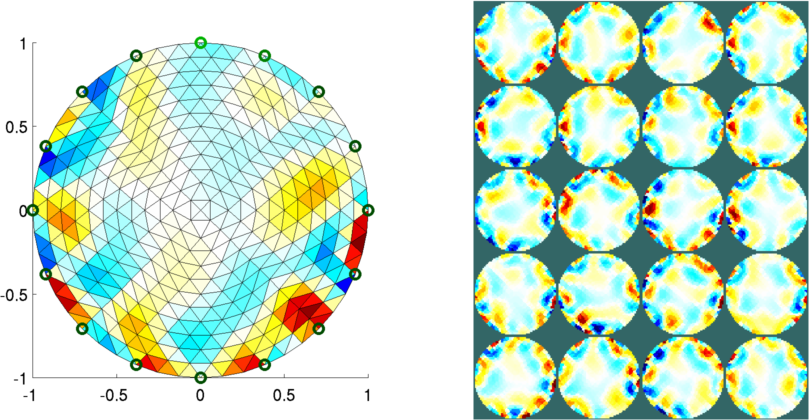
Figure: Reconstructed images of targets moving in the mesh Left Reconstruction mesh with first image Right Reconstructed images of all targets Exploring the effect of mesh densityThis image shows a large amount of what looks like noise. In order to explore the effect of mesh density and electrode refinement, we modify the parameters n_nodes and refine_level (electrode refinement.Mesh with n_nodes=500 and refine_level=2
Figure: Animation of the simulation mesh surrounding a moving target 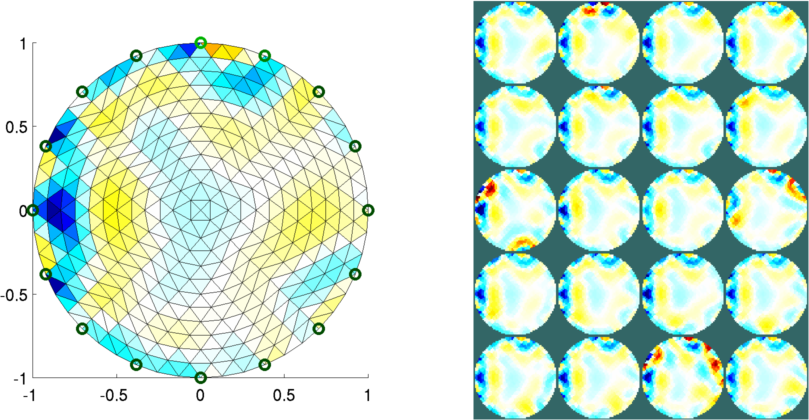
Figure: Reconstructed images of targets moving in the mesh Left Reconstruction mesh with first image Right Reconstructed images of all targets Mesh with n_nodes=1000 and refine_level=4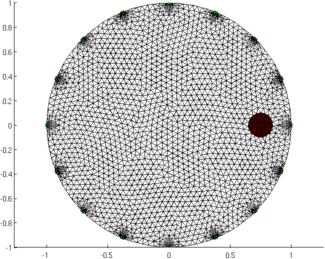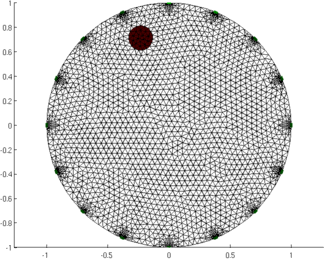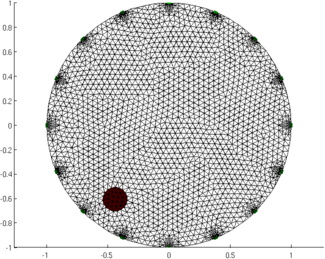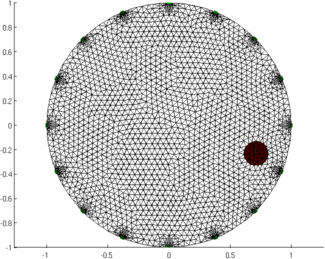
Figure: Animation of the simulation mesh surrounding a moving target 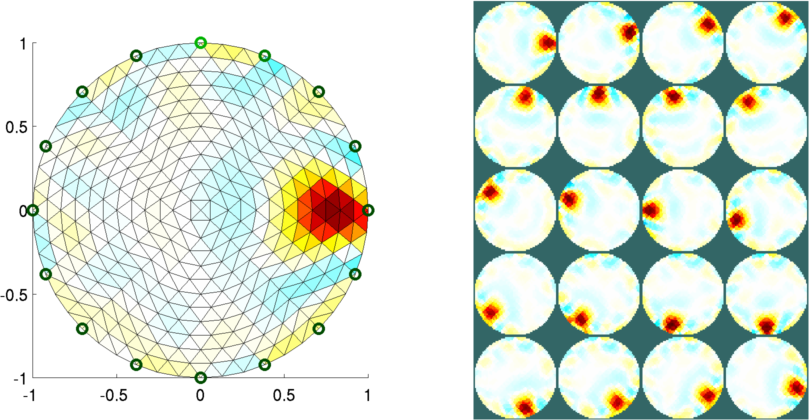
Figure: Reconstructed images of targets moving in the mesh Left Reconstruction mesh with first image Right Reconstructed images of all targets |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $