

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Dual Models to reconstruct a slice of a volumeA common use of dual models is to allow the forward model to represent the entire space, while the reconstruction model represents a slice through the volume at the place of interest.To simulate this, we simulate a ball moving in a helical path on a fine netgen model.
% Simulate Moving Ball - Helix $Id: centre_slice01.m 2666 2011-07-12 20:41:02Z aadler $
% get ng_mdl_16x2_vfine from data_contrib section of web page
n_sims= 20;
stim = mk_stim_patterns(16,2,'{ad}','{ad}',{},1);
fmdl = mk_library_model('cylinder_16x2el_vfine');
fmdl.stimulation = stim;
[vh,vi,xyzr_pt]= simulate_3d_movement( n_sims, fmdl);
clf; show_fem(fmdl)
crop_model(gca, inline('x-z<-15','x','y','z'))
view(-23,14)
hold on
[xs,ys,zs]=sphere(10);
for i=1:n_sims
xp=xyzr_pt(1,i); yp=xyzr_pt(2,i);
zp=xyzr_pt(3,i); rp=xyzr_pt(4,i);
hh=surf(rp*xs+xp, rp*ys+yp, rp*zs+zp);
set(hh,'EdgeColor',[.4,0,.4],'FaceColor',[.2,0,.2]);
end
hold off
print_convert centre_slice01a.png '-density 100'
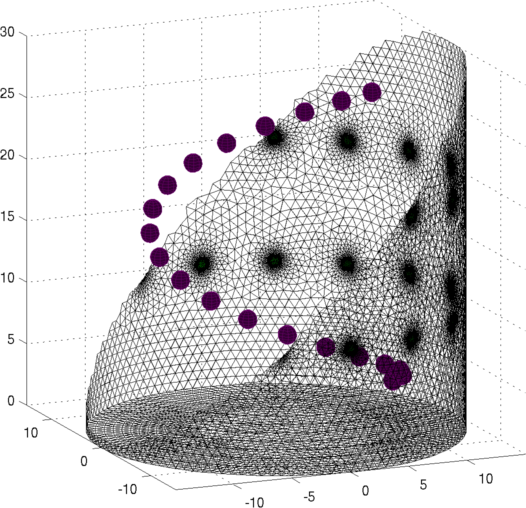
Figure: Netgen model of a 2×16 electrode tank. The positions of the simulated conductive target moving in a helical path are shown in purple. Create reconstruction modelIn order to reconstruct the image, we use a dual model where the 2D coarse model is mapped to only a layer of elements in the fine model.
% 2D solver $Id: centre_slice02.m 2666 2011-07-12 20:41:02Z aadler $
% Create and show inverse solver
imdl = mk_common_model('b3cr',[16,2]);
f_mdl = mk_library_model('cylinder_16x2el_coarse');
f_mdl.stimulation = stim;
imdl.fwd_model = f_mdl;
% Create coarse model
imdl2d= mk_common_model('b2c2',16);
c_mdl= imdl2d.fwd_model;
% Show fine model
show_fem(f_mdl);
crop_model(gca, inline('x-z<-15','x','y','z'))
view(-23,10)
scl= 15; % scale difference between c_mdl and f_mdl
c_els= c_mdl.elems;
c_ndsx= c_mdl.nodes(:,1)*scl;
c_ndsy= c_mdl.nodes(:,2)*scl;
c_ndsz= 0*c_ndsx;
layersep= .3;
layerhig= .1;
hold on
% Lower resonstruction layer
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(1-layersep)*scl);
set(hh, 'EdgeColor', [1,0,0]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(1-layersep-layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(1-layersep+layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
% Upper resonstruction layer
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(1+layersep)*scl);
set(hh, 'EdgeColor', [0,0,1]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(1+layersep-layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(1+layersep+layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
for i=1:n_sims
xp=xyzr_pt(1,i); yp=xyzr_pt(2,i);
zp=xyzr_pt(3,i); rp=xyzr_pt(4,i);
hh=surf(rp*xs+xp, rp*ys+yp, rp*zs+zp);
set(hh,'EdgeColor',[.4,0,.4]);
end
hold off;
print_convert centre_slice02a.png '-density 100'
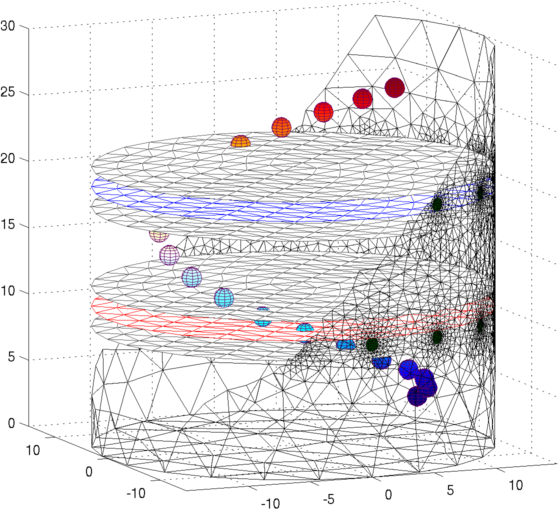
Figure: Netgen model of a 2×16 electrode tank. The positions of the simulated conductive target moving in a helical path are shown in purple. The 3D fine model is shown (cropped). The upper (blue) and lower (red) layers corresponding to the geometry of the coarse model are shown. The z direction limits of the coarse model are shown in grey. Image reconstructionsFirst, we create a coarse model which represents the entire depth in z (ie. like the 2½D model). Images reconstructed with this model have more artefacts, but show the reconstructed target at all depths.% 2D solver $Id: centre_slice03.m 4839 2015-03-30 07:44:50Z aadler $ % Set coarse as reconstruction model imdl.rec_model= c_mdl; c_mdl.mk_coarse_fine_mapping.f2c_offset = [0,0,scl]; c_mdl.mk_coarse_fine_mapping.f2c_project = (1/scl)*speye(3); c_mdl.mk_coarse_fine_mapping.z_depth = inf; c2f= mk_coarse_fine_mapping( f_mdl, c_mdl); imdl.fwd_model.coarse2fine = c2f; imdl.RtR_prior = @prior_gaussian_HPF; imdl.solve = @inv_solve_diff_GN_one_step; imdl.hyperparameter.value= 0.1; imgc= inv_solve(imdl, vh, vi); clf; show_slices(imgc); print_convert centre_slice04a.png '-density 75';Next, we create coarse models which represent the a thin 0.1×scale slice in z. These images display a targets in the space from the original volume. % 2D solver $Id: centre_slice04.m 4839 2015-03-30 07:44:50Z aadler $ imdl.hyperparameter.value= 0.05; c_mdl.mk_coarse_fine_mapping.f2c_offset = [0,0,(1-.3)*scl]; c_mdl.mk_coarse_fine_mapping.z_depth = 0.1; c2f= mk_coarse_fine_mapping( f_mdl, c_mdl); imdl.fwd_model.coarse2fine = c2f; imgc0= inv_solve(imdl, vh, vi); % Show image of reconstruction in upper planes show_slices(imgc0); print_convert centre_slice04b.png '-density 75'; c_mdl.mk_coarse_fine_mapping.f2c_offset = [0,0,(1+.3)*scl]; c_mdl.mk_coarse_fine_mapping.z_depth = 0.1; c2f= mk_coarse_fine_mapping( f_mdl, c_mdl); imdl.fwd_model.coarse2fine = c2f; imgc1= inv_solve(imdl, vh, vi); % Show image of reconstruction in lower planes show_slices(imgc1); print_convert centre_slice04c.png '-density 75'; 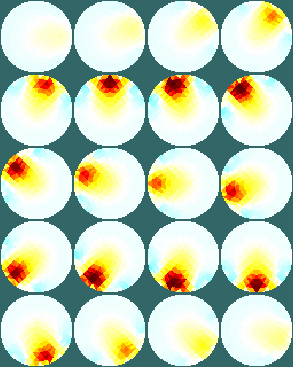
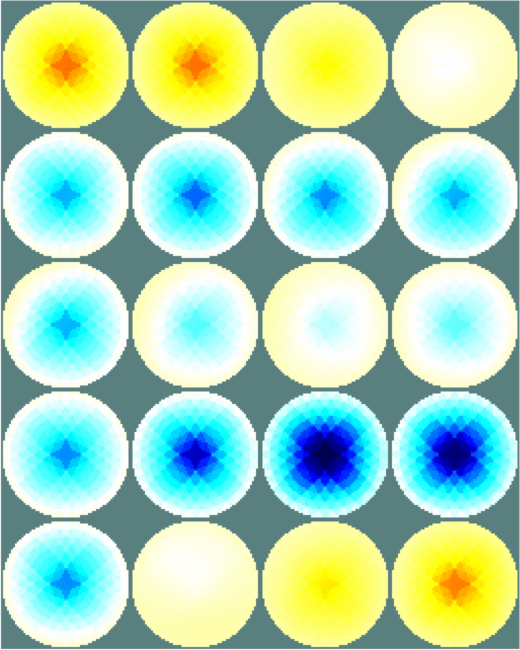
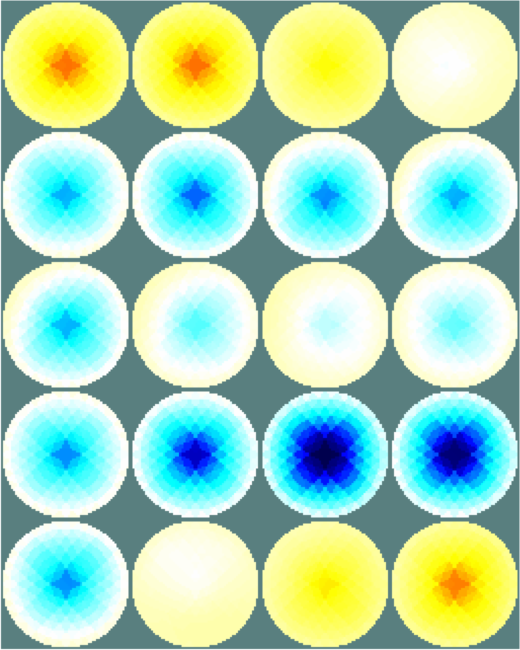
Figure: Reconstructed images of a target moving in a helical pattern using difference coarse models Left coarse model with zdepth=∞ Centrecoarse model with zdepth=0.1×scale at upper position Right coarse model with zdepth=0.1×scale at lower positions Simpler reconstruction modelThe previous model requires lots of time an memory to calculate the Jacobian for the reconstruction, because of the large number of FEMs. To speed up the calcualion, we use a simpler fine model.
% 2D solver $Id: centre_slice05.m 2673 2011-07-13 06:57:41Z aadler $
% Create and show inverse solver
imdl = mk_common_model('b3cr',[16,2]);
f_mdl= imdl.fwd_model;
% Create coarse model
imdl2d= mk_common_model('b2c2',16);
c_mdl= imdl2d.fwd_model;
% Show fine model
clf;show_fem(f_mdl);
crop_model(gca, inline('x-z<-.5','x','y','z'))
view(-23,10)
scl= 1; % scale difference between c_mdl and f_mdl
c_els= c_mdl.elems;
c_ndsx= c_mdl.nodes(:,1)*scl;
c_ndsy= c_mdl.nodes(:,2)*scl;
c_ndsz= 0*c_ndsx;
layersep= .3;
layerhig= .1;
hold on
% Lower resonstruction layer
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(-layersep)*scl);
set(hh, 'EdgeColor', [1,0,0]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(-layersep-layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(-layersep+layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
% Upper resonstruction layer
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(+layersep)*scl);
set(hh, 'EdgeColor', [0,0,1]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(+layersep-layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
hh= trimesh(c_els, c_ndsx, c_ndsy, c_ndsz+(+layersep+layerhig)*scl);
set(hh, 'EdgeColor', [.3,.3,.3]);
[xs,ys,zs]=sphere(10);
for i=1:size(xyzr_pt,2)
xp=xyzr_pt(1,i)/15; yp=xyzr_pt(2,i)/15;
zp=xyzr_pt(3,i)/15-1; rp=xyzr_pt(4,i)/15;
hh=surf(rp*xs+xp, rp*ys+yp, rp*zs+zp);
set(hh,'EdgeColor',[.4,0,.4]);
end
hold off;
print_convert centre_slice05a.png '-density 100';
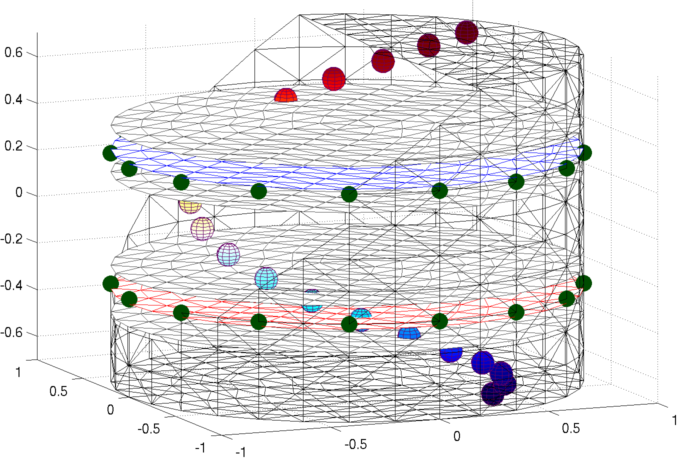
Figure: Simple extruded model of a 2×16 electrode tank. The positions of the simulated conductive target moving in a helical path are shown in purple. The 3D fine model is shown (cropped). The upper (blue) and lower (red) layers corresponding to the geometry of the coarse model are shown. The z direction limits of the coarse model are shown in grey. Image reconstructionsWe reconstruct with coarse models with a) the entire depth in z (ie. like the 2½D model). Images reconstructed with this model have more artefacts, b) we create coarse models which represent the a thin 0.1×scale slice in z. These images display a targets in the space from the original volume.% 2D solver $Id: centre_slice06.m 4839 2015-03-30 07:44:50Z aadler $ % Set coarse as reconstruction model imdl.rec_model= c_mdl; c2f= mk_coarse_fine_mapping( f_mdl, c_mdl); imdl.fwd_model.coarse2fine = c2f; imdl.RtR_prior = @prior_gaussian_HPF; imdl.solve = @inv_solve_diff_GN_one_step; imdl.hyperparameter.value= 0.2; imgc= inv_solve(imdl, vh, vi); clf; show_slices(imgc); print_convert centre_slice06a.png '-density 75'; c_mdl.mk_coarse_fine_mapping.f2c_offset = [0,0,-.3]; c_mdl.mk_coarse_fine_mapping.z_depth = 0.1; c2f= mk_coarse_fine_mapping( f_mdl, c_mdl); imdl.fwd_model.coarse2fine = c2f; imgc0= inv_solve(imdl, vh, vi); imgc0= inv_solve(imdl, vh, vi); show_slices(imgc0); print_convert centre_slice06b.png '-density 75'; c_mdl.mk_coarse_fine_mapping.f2c_offset = [0,0,.3]; c_mdl.mk_coarse_fine_mapping.z_depth = 0.1; c2f= mk_coarse_fine_mapping( f_mdl, c_mdl); imdl.fwd_model.coarse2fine = c2f; imgc1= inv_solve(imdl, vh, vi); show_slices(imgc1); print_convert centre_slice06c.png '-density 75'; 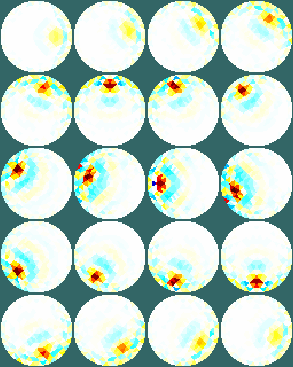
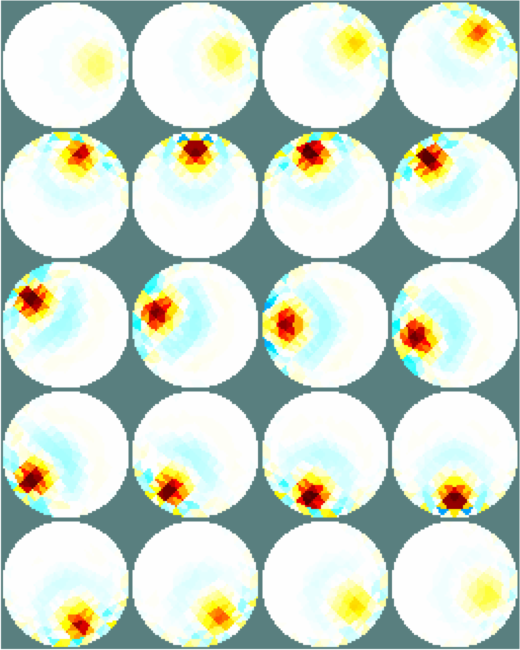
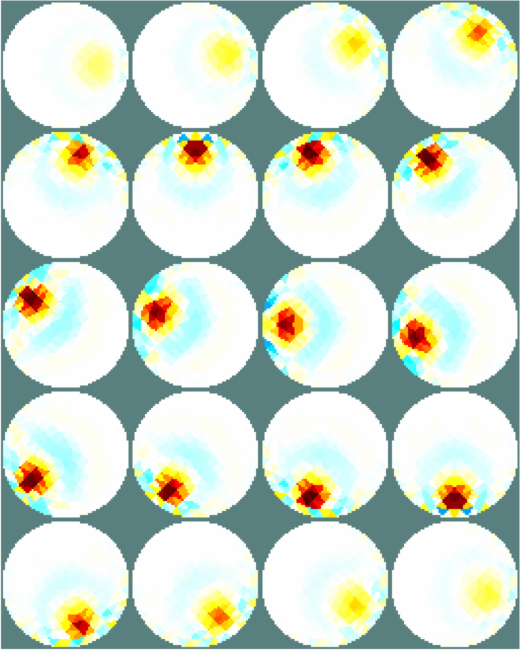
Figure: Reconstructed images of a target moving in a helical pattern using difference coarse models Left coarse model with zdepth=∞ Centrecoarse model with zdepth=0.1×scale at upper position Right coarse model with zdepth=0.1×scale at lower positions |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $