

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Dual Models − Coarse model partially covers fine modelIn many cases, the inverse model only covers part of the space. It is necessary to cover the entire space with the fine model to provide an accurate forward solution, but it is not possible to accurately calculate parameters for the low sensitivity parts of the space.Coarse and Fine modelsCreate a fine model of a tank with 7 electrodes just near one side. The coarse model will only represent the conductivity near this region.
% Dual Partial $Id: dual_partial2d01.m 3504 2012-07-05 11:53:21Z aadler $
imdl= mk_common_model('d2c2',64);
fmdl= imdl.fwd_model;
% Only keep 11 electrodes on top
fmdl.electrode = fmdl.electrode([60:64,1:6]);
% New stimulation pattern with 11 electrodes
fmdl.stimulation = mk_stim_patterns(11,1,'{ad}','{ad}',{},1);
% Remove meas_select - it was created for 64 electrodes
fmdl = rmfield(fmdl,'meas_select');
subplot(121)
show_fem(fmdl); axis square
% Crop model
[cmdl,c2f_idx]= crop_model(fmdl, inline('(y-0.25)-abs(x)<0','x','y','z'));
subplot(122)
show_fem(cmdl);
axis(1.05*[-1 1 -1 1]); axis square;
print_convert dual_partial2d01a.png;
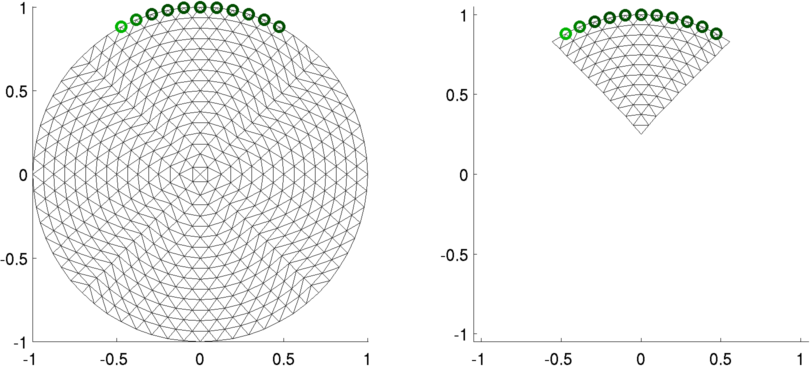
Figure: Fine (left) and dual (right) models Simulated dataData are simulated of a point rotating on a fine mesh (inverse crime in this case)
% Simulate object moving $Id: dual_partial2d02.m 2782 2011-07-14 21:14:12Z aadler $
% simulation model
imdl= mk_common_model('d2c2',64);
smdl= imdl.fwd_model;
% Only keep 11 electrodes on top
smdl.electrode = smdl.electrode([60:64,1:6]);
smdl.stimulation = mk_stim_patterns(11,1,'{ad}','{ad}',{},1);
[vh,vi,xyr]= simulate_2d_movement(32, smdl, [0.75,0.05] );
% Only 12 to 2 O'clock
idx= 5:10;
vi= vi(:,idx); xyr= xyr(:,idx);
clf;subplot(121)
show_fem(smdl); axis square
% Show target positions
theta= linspace(0,2*pi,50); xr= cos(theta); yr= sin(theta);
hold on;
for i=1:size(xyr,2)
hh= plot(xyr(3,i)*xr+ xyr(1,i),xyr(3,i)*yr+ xyr(2,i));
set(hh,'LineWidth',3,'Color',[0,0,0]);
end
hold off;
print_convert dual_partial2d02a.png;

Figure: Simulation data Reconstruction modelsCreate fine mesh (fmdl_mdl) and dual mesh (dmdl_mdl) reconstruction models.% Dual Partial $Id: dual_partial2d03.m 4839 2015-03-30 07:44:50Z aadler $ % base model imdl.RtR_prior = @prior_noser; imdl.hyperparameter.value = .2; % Reconstruction model - only fine reconstruction frec_mdl= imdl; frec_mdl.fwd_model= fmdl; % fine model % Reconstruction model - dual model drec_mdl = imdl; drec_mdl.fwd_model= fmdl; % fine model % coarse to fine mapping nf_els= size(fmdl.elems,1); nc_els= size(cmdl.elems,1); drec_mdl.fwd_model.coarse2fine= sparse(c2f_idx, 1:nc_els, 1, nf_els, nc_els); Reconstructed images (projected to fine mesh)% Dual Partial $Id: dual_partial2d04.m 2749 2011-07-14 13:32:13Z aadler $ clf; levels= [inf,inf,0,1,1]; % reconstruct fine model imgf= inv_solve(frec_mdl, vh, vi); show_slices(imgf,levels); print_convert dual_partial2d04a.png; % reconstruct dual model imgd= inv_solve(drec_mdl, vh, vi); show_slices(imgd,levels); print_convert dual_partial2d04b.png; 

Figure: Fine (top) and dual (bottom) reconstructed images Reconstructed images (projected to coarse mesh)% Dual Partial $Id: dual_partial2d05.m 2749 2011-07-14 13:32:13Z aadler $ clf; levels= [inf,inf,0,1,1]; % reconstruct fine model params onto coarse model imgf= inv_solve(frec_mdl, vh, vi); imgf.fwd_model= cmdl; imgf.elem_data= imgf.elem_data(c2f_idx,:); show_slices(imgf,levels); print_convert dual_partial2d05a.png; % reconstruct dual model on coarse mesh imgd= inv_solve(drec_mdl, vh, vi); imgd.fwd_model= cmdl; show_slices(imgd,levels); print_convert dual_partial2d05b.png; 
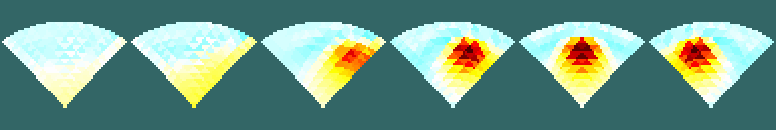
Figure: Fine (top) and dual (bottom) reconstructed images |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $