

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
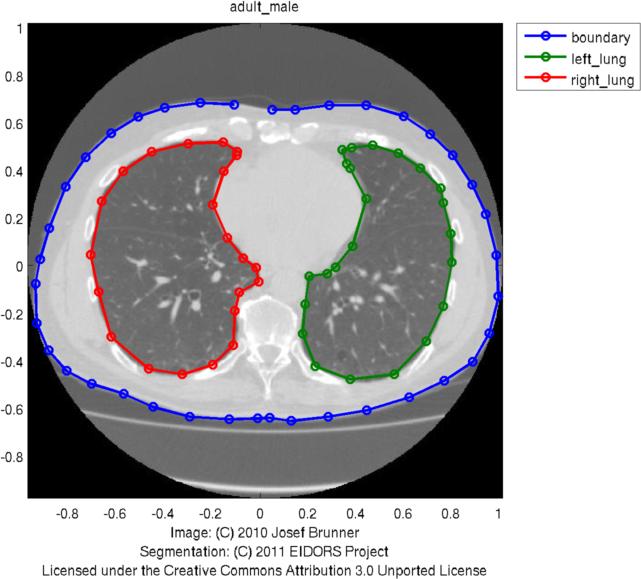
Modeling EIT current flow in a human thorax modelThis example shows how EIDORS can by use Netgen to model the body shape of a human and then use it to build a reconstruction algorithm and see the current flow in the body. For this exmample, you need at least Netgen version 4.9.13. These images are designed to be used on the Wikipedia EIT page. All images on this page are licenced under the Creative Commons Attribution License.Load thorax shape and identify contoursThis image is from a human CT of a healthy man, provided with permission. Hand registered contours are available in the eidors shape_library. The image is available here [thorax-mdl.jpg]
% get contours
thorax = shape_library('get','adult_male','boundary');
rlung = shape_library('get','adult_male','right_lung');
llung = shape_library('get','adult_male','left_lung');
% one could also run:
% shape_library('get','adult_male');
% to get all the info at once in a struct
% show the library image
shape_library('show','adult_male');
print_convert thoraxmdl01a.jpg '-density 100'
Use Netgen to build a FEM model of the thorax
shape = { 1, % height
{thorax, rlung, llung}, % contours
[4,50], % perform smoothing with 50 points
0.04}; % small maxh (fine mesh)
elec_pos = [ 16, % number of elecs per plane
1, % equidistant spacing
0.5]'; % a single z-plane
elec_shape = [0.05, % radius
0, % circular electrode
0.01 ]'; % maxh (electrode refinement)
fmdl = ng_mk_extruded_model(shape, elec_pos, elec_shape);
% this similar model is also available as:
% fmdl = mk_library_model('adult_male_16el_lungs');
[stim,meas_sel] = mk_stim_patterns(16,1,[0,1],[0,1],{'no_meas_current'}, 1);
fmdl.stimulation = stim;
img=mk_image(fmdl,1);
img.elem_data(fmdl.mat_idx{2})= 0.3; % rlung
img.elem_data(fmdl.mat_idx{3})= 0.3; % llung
clf; show_fem(img); view(0,70);
print_convert thoraxmdl02a.jpg '-density 100'

Voltage and Current Distribution in a sliceVoltage distributionimg_v = img; % Stimulate between elecs 16 and 5 to get more interesting pattern img_v.fwd_model.stimulation(1).stim_pattern = sparse([16;5],1,[1,-1],16,1); img_v.fwd_solve.get_all_meas = 1; vh = fwd_solve(img_v); img_v = rmfield(img, 'elem_data'); img_v.node_data = vh.volt(:,1); img_v.calc_colours.npoints = 128; PLANE= [inf,inf,0.35]; % show voltages on this slice subplot(221); show_slices(img_v,PLANE); axis off; axis equal print_convert thoraxmdl03a.jpg %Current distribution img_v = img; img_v.fwd_model.mdl_slice_mapper.npx = 64; img_v.fwd_model.mdl_slice_mapper.npy = 64; img_v.fwd_model.mdl_slice_mapper.level = PLANE; q = show_current(img_v, vh.volt(:,1)); quiver(q.xp,q.yp, q.xc,q.yc,10,'b'); axis tight; axis image; ylim([-1 1]);axis off print_convert thoraxmdl04a.jpg  
Left Voltage distribution and Right Current distribution in slices at z= 0.35. Current streamlines and the original image
img_v.fwd_model.mdl_slice_mapper.npx = 1000;
img_v.fwd_model.mdl_slice_mapper.npy = 1000;
img_v.fwd_model.mdl_slice_mapper.level = PLANE;
% Calculate at high resolution
q = show_current(img_v, vh.volt(:,1));
pic = shape_library('get','adult_male','pic');
imagesc(pic.X, pic.Y, pic.img);
% imgt= flipdim(imread('thorax-mdl.jpg'),1); imagesc(imgt);
colormap(gray(256)); set(gca,'YDir','normal');
hold on
sx = linspace(-.5,.5,15)';
sy = 0.05 + linspace(-.5,.5,15)';
hh=streamline(q.xp,q.yp, q.xc, q.yc,sx,sy); set(hh,'Linewidth',2, 'color','b');
hh=streamline(q.xp,q.yp,-q.xc,-q.yc,sx,sy); set(hh,'Linewidth',2, 'color','b');
axis equal; axis tight; axis off; print_convert thoraxmdl05a.jpg
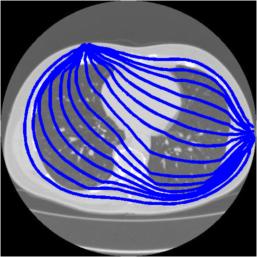
Stream lines through z= 0.35. The path of the streamslines is limited by the density of the underlying FEM. With a finer mesh FEM, it would be possible to calculate streamlines that do not display path irregularities. Add Equipotential lines to the original image
img_v = img;
% Stimulate between elecs 16 and 5 to get more interesting pattern
img_v.fwd_model.stimulation(1).stim_pattern = sparse([16;5],1,[1,-1],16,1);
img_v.fwd_solve.get_all_meas = 1;
vh = fwd_solve(img_v);
img_v = rmfield(img, 'elem_data');
img_v.node_data = vh.volt(:,1);
img_v.calc_colours.npoints = 256;
imgs = calc_slices(img_v,PLANE);
clf
imagesc(pic.X, pic.Y, pic.img); colormap(gray(256)); set(gca,'YDir','normal');
hh=streamline(q.xp,q.yp, q.xc, q.yc,sx,sy); set(hh,'Linewidth',2);
hh=streamline(q.xp,q.yp,-q.xc,-q.yc,sx,sy); set(hh,'Linewidth',2);
[x y] = meshgrid( linspace(pic.X(1), pic.X(2),size(imgs,1)), ...
linspace(pic.Y(2), pic.Y(1),size(imgs,2)));
hold on;
contour(x,y,imgs,31);
hh= findobj('Type','patch'); set(hh,'LineWidth',2)
hold off; axis off; axis equal; %ylim([50,450]);
print_convert thoraxmdl06a.jpg
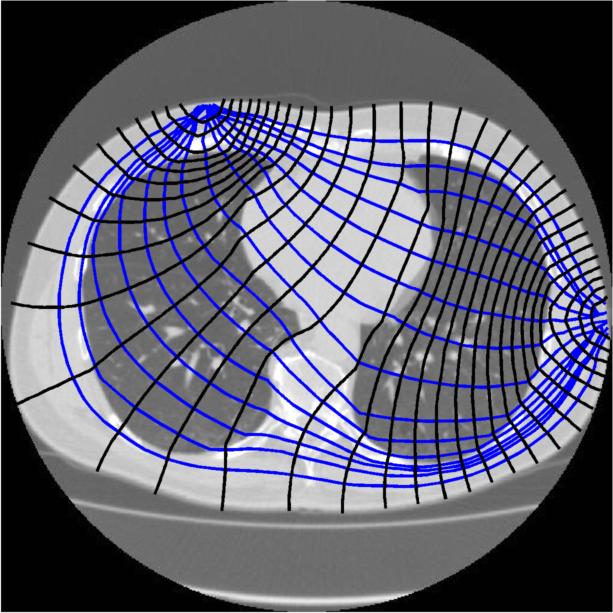
Stream lines and equipotiential through z= 0.35. |
Last Modified: $Date: 2017-02-28 13:21:02 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $